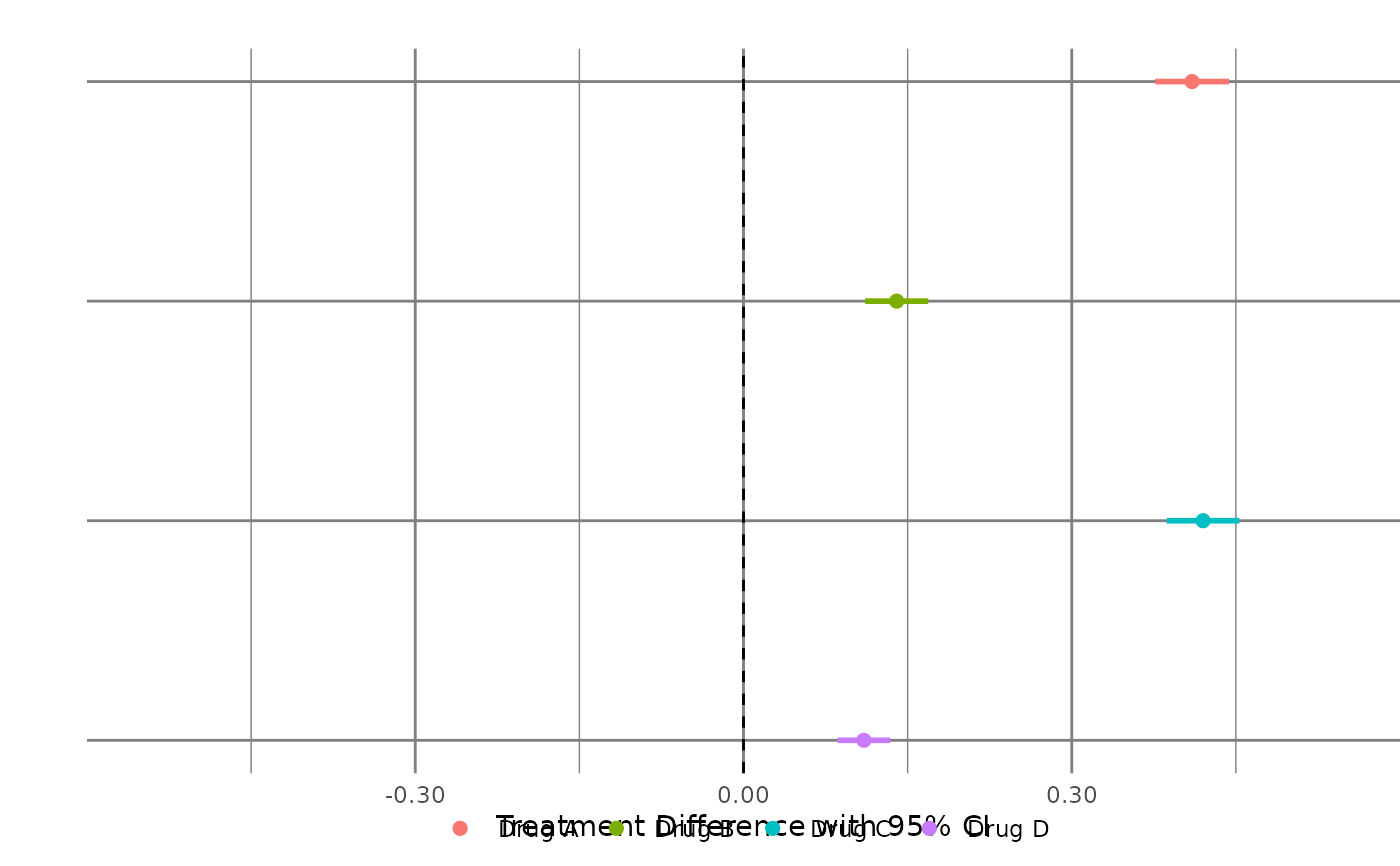
Create Forest plot
generate_fig_rft(
data,
xlabel = NULL,
ylabel = NULL,
fact_subset = "Benefit",
type_subset = "Binary",
type_scale = "Fixed",
scale_x = "Identity",
x_scale_n1_p1 = "N",
select_nnx
)Arguments
- data
(
character) Data for figure- xlabel
(
character) Label for x-axis- ylabel
(
character) Label for y-axis- fact_subset
(
character) filter data- type_subset
(
character) Selected subset "Binary" or "Continuous"- type_scale
(
character) selected scale display type- scale_x
(
character) Type of scale for x-axis "Identity" or "log10"- x_scale_n1_p1
(
character) fix x-axis scale between -1 and 1- select_nnx
(
character) show NNT/NNH
Examples
dot_plot_src <- subset(effects_table, !is.na(Prop1))
bdin <- subset(dot_plot_src, Factor == "Benefit")
rdin <- subset(dot_plot_src, Factor == "Risk")
fplot_data <- prepare_dot_forest_plot_data(
data = dot_plot_src,
drug = unique(dot_plot_src$Trt1),
benefit = unique(bdin$Outcome),
risk = unique(rdin$Outcome),
filters = "None",
category = "All",
type_graph = "Absolute risk",
type_risk = "Crude proportions",
ci_method = "Calculated"
)
#> [2025-02-12 02:27:14] > Prepare Dot plot data for binary outcomes
#> [2025-02-12 02:27:14] > trigger analysis based on type
#> [2025-02-12 02:27:14] >
#> absolute risk CI for binary outcomes is calculated and saved
#> [2025-02-12 02:27:14] > Prepare Forest plot data for absolute risk
#> [2025-02-12 02:27:14] > Prepare Dot plot data for binary outcomes
#> [2025-02-12 02:27:14] > trigger analysis based on type
#> [2025-02-12 02:27:14] >
#> absolute risk CI for binary outcomes is calculated and saved
#> [2025-02-12 02:27:14] > Prepare Forest plot data for absolute risk
#> [2025-02-12 02:27:14] > Dataout object from the create_order_label_der function is created
#> [2025-02-12 02:27:14] > Prepare data for Dot and Forest plots
forest_plot_data <- subset(
fplot_data$forest_plot_data,
factor == "Benefit" & type == "Binary"
)
generate_fig_rft(
data = forest_plot_data,
fact_subset = "Benefit",
type_subset = "Binary",
xlabel = "Treatment Difference with 95% CI\n",
select_nnx = "No"
)
#> Warning: Unknown or uninitialised column: `Trt2`.
#> [2025-02-12 02:27:14] > Create Forest plot